Information retrieval on CORD-19 papers about COVID-19 Part 2: Information Retrieval Using Elasticsearch
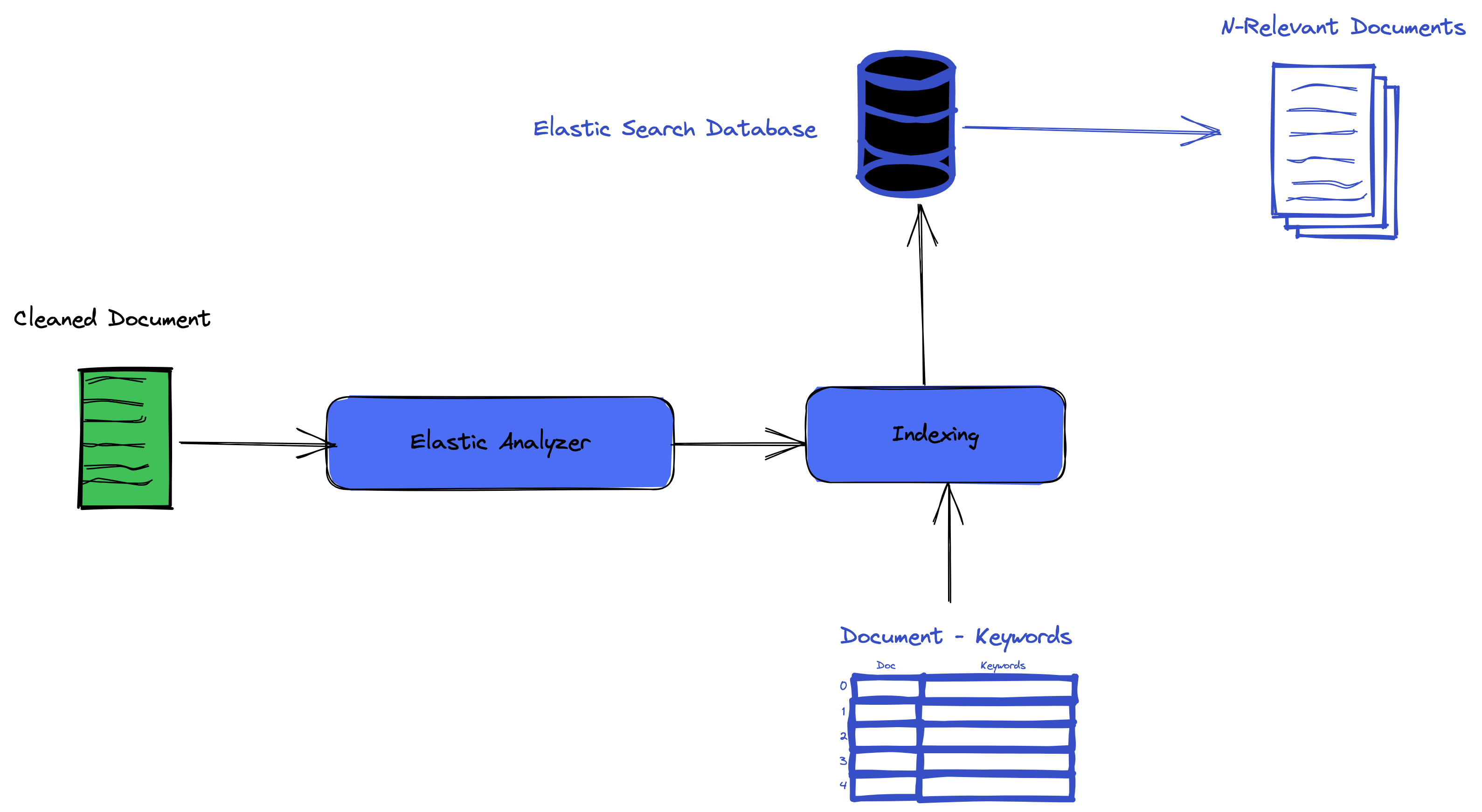
In part one of this tutorial, we cleaned our dataset and searched using TF-IDF similarity. In this part, we will learn how to add our documents in Elastic-search, index them, and query our index. By the end of this second part of this series, you should have a complete working system you can use to make queries to Elasticsearch.
To run this second part, make sure you have :
- Make sure to have the dataset with cleaned abstract
- A mapping of the document pub medical id and the keyword for each document.
-
Elasticsearch is installed and running in your system; if you don’t have it installed, you can follow this tutorial to install it in your system.
- You should also have the elasticsearch-dsl libray installed in your python environment; it will help us to run our queries and interact with the Elasticsearch database.
Different ways to interact with Elasticsearch

In this section, we will discuss different ways to interact with Elasticsearch, and at the end, we will confirm if we can connect to it from our machine.
In the literature, there are different ways to interact with the Elasticsearch database, but in general, any tools that can help you to make an HTTP request with a JSON body can do the job.
The most important part of this is understanding how the interaction is done and that we are good to go. The tool we are using to make the request is pint-size. In general, the interaction with Elasticsearch is done via an HTTP request. In the literature, there are different ways to interact with the Elasticsearch database, but in general, any tools that can help you to make an HTTP request with a JSON body can do the job.
Among the tools you will find out there there are :
- Kibana: I have never used it before, but it looks like a dashboard that can help you make your request, format your JSON data and visualize your data.
- Since we are making requests, you can use the
curlcommand from Linux to interact with Elasticsearch via the command line. - Postman: For those who have worked and tested Restful APIs before, this tool can come to your rescue.
- Any low-level library from a programming language that can help make HTTP requests. In this category comes the python-elasticsearch package. A far high-level library built on top of the low-level library offers a far more convenient way to connect with Elasticsearch. I prefer this because of my software engineering background and since I have worked with Object Relational Mapping before. This library is similar to a Relational Object Framework because it provides an optional wrapper for working with documents as Python objects: defining mappings, retrieving and saving documents, wrapping the document data in user-defined classes. Just like SQL Alchemy if you are familiar with ORM in python. For the rest of the tutorial, we will be using this high-level library to interact with Elasticsearch. In some other circumstances, we will use the low-level API.
To check if Elasticsearch is installed and working, we can run the following code in your command line:
curl --location --request GET 'localhost:9200'
If everything is running and installed, you should see the following output.
{
"name" : "your-local-macadress.local",
"cluster_name" : "elasticsearch_es.py",
"cluster_uuid" : "t__AU_HQTtiUSsI-wEe_Gw",
"version" : {
"number" : "7.16.3",
"build_flavor" : "default",
"build_type" : "tar",
"build_hash" : "4e6e4eab2297e949ec994e688dad46290d018022",
"build_date" : "2022-01-06T23:43:02.825887787Z",
"build_snapshot" : false,
"lucene_version" : "8.10.1",
"minimum_wire_compatibility_version" : "6.8.0",
"minimum_index_compatibility_version" : "6.0.0-beta1"
},
"tagline": "You Know, for Search."
}
If you are getting any errors, check if your installation was successful or google the error message. I am sure you can find an answer from the first google search response.
With our Elasticsearch running, let us build the model to handle our data.
Building the Document Model.
Let’s recall a model from an ORM perspective. A model is mapping a class in a programming language to a table in a Query language. The exact definition is valid for Elasticsearch. In this case, a model is a mapping of a class in python to an index in elastic search.
Analyzer
An analyzer is a function used to preprocess the text when it is saved to Elasticsearch or querying a field in elastic search.
It contains three lower-level building blocks: character filters, tokenizers, and token filters.
This can help preprocess the text as we did in the first section but on the Elasticsearch level.
I am not sure if we need an analyzer after preprocessing our text with python but let us do it again for the demonstration process.
from elasticsearch_dsl import analyzer, tokenizer, connections
article_analyzer = analyzer('analyzer',
tokenizer=tokenizer('trigram', 'nGram', min_gram=2, max_gram=3),
stopwords=['_english_'], filter=['lowercase', "asciifolding"])
As you can see this analyzer anyttime we are applying it to a field in our index it will remove english stopwords , filter ngrams and only keep bigrams and trigrams , and remove non ascii characters.
A lot have been done already on the preprocessing text, but we could also do the stemmming and all the cleaning process direcly via the analyzer in ElasticSearch.
connections.create_connection(hosts=['localhost'])
<Elasticsearch([{'host': 'localhost'}])>
from elasticsearch_dsl import Document,Date,Integer,Keyword,Text, connections,AttrList,Nested,Float,InnerDoc
from elasticsearch.helpers import bulk
class Article(Document):
title = Text(analyzer=article_analyzer, term_vector="with_positions_offsets_payloads")
abstract = Text(analyzer=article_analyzer, term_vector="with_positions_offsets_payloads", store=True)
tags = Keyword(multi=True)
publish_time = Date()
authors = Keyword(multi=True)
class Index:
name = 'covid19'
settings = {"number_of_shards": 2, "mapping":{"total_fields": {"limit": 10000}}}
def add_tag(self, tag):
self.tags.append(tag)
def save(self, ** kwargs):
return super(Article, self).save(** kwargs)
@classmethod
def bulk_save(cls, dataframe, document_tfidf):
"""
this should take a dataframe and save all it content to elasticsearch
"""
articles = []
for index, row in dataframe.iterrows():
tags = list(document_tfidf.get(index).keys())
article = cls(meta={'id': index},
title=row['title'],
abstract=row['abstract_cleaned'],
publish_time=row['publish_time'],
authors=row['authors'])
for tag in tags:
article.add_tag(tag)
articles.append(article.to_dict(include_meta=True))
for i in range(0, len(articles), 100):
client = connections.get_connection()
bulk(client, articles[i:i+100], request_timeout=120 * 10 )
print("done saving for the {}th batch".format(i))
This will be our model , let us add the attribute to it :
title = Text(analyzer=article_analyzer)
The above line creates the title and adds the analyzer to it. We also specify the data type and say that it is a text.
abstract = Text(analyzer=article_analyzer)
The same applies to the abstract.
For the next attribute the tags we have the following :
tags = Keyword(multi=True)
This says that it is a keyword and can have a list of values hence the parameter multi.
To save the publish date of an article, we have the following :
publish_time = Date()
It says that our publishing time is a date.
To save the article’s authors, we use the same approach as for the tags.
authors = Keyword(multi=True)
With all our fields declared, let’s move to the configuration for the class.
This is done by adding a metadata class.
class Index:
name = 'covid19'
settings = {"number_of_shards": 2, "mapping":{"total_fields": {"limit": 10000}}}
This specifies the index name to use and other configuration settings.
Next, we need a method to save our articles :
Next, we have a bulk_save method, and this will take our dataframe, the mapping of an article, and keyword and loop over it and save the article in a batch of 100 in our index.
This is not the best optimal way to do this; I am sure I will be looking for a faster way to do this using Elastisearch and dask.
With our model defined, and utility methods described, we can create the index in the database.
Before we first use the Article document type, we need to create the mappings in Elasticsearch. For that, we can either use the Index object or make the mappings directly by calling the init class method:
from elasticsearch_dsl import Index
old_index = Index('covid19')
try:
old_index.delete()
except Exception:
pass
Article.init()
None
The above code will create the index, it is similar to running the migration when we are working with object relational mappers.
If there was no index named covid19 in our database , it will create the index for us.
If the index exist it will throw an error we will have to delete the index manualy and rerun the line.
To delete the index , run the following command in your cmd:
curl --location --request DELETE 'localhost:9200/covid19'
If everything goes well we should have our index structured in the database. To check if the index is there and the mapping are done correctly , run the following command :
curl --location --request GET 'localhost:9200/covid19/_mapping'
This will return the mapping if everything was done correctly .
{
"covid19": {
"mappings": {
"properties": {
"abstract": {
"type": "text",
"analyzer": "analyzer"
},
"authors": {
"type": "keyword"
},
"publish_time": {
"type": "date"
},
"tags": {
"type": "keyword"
},
"title": {
"type": "text",
"analyzer": "analyzer"
}
}
}
}
}
Let us run the code to save our dataset in the index :
Article.bulk_save(data_df, document_tfidf)
done saving for the 0th batch
done saving for the 100th batch
done saving for the 200th batch
done saving for the 300th batch
done saving for the 400th batch
done saving for the 500th batch
done saving for the 600th batch
done saving for the 700th batch
done saving for the 800th batch
done saving for the 900th batch
done saving for the 1000th batch
This will go over our dataframe and save each document in our index.
It can take sometime but once it is done we will have our document indexed in elasticsearch.
We can now move to the final section of this serie , querying .
Querying
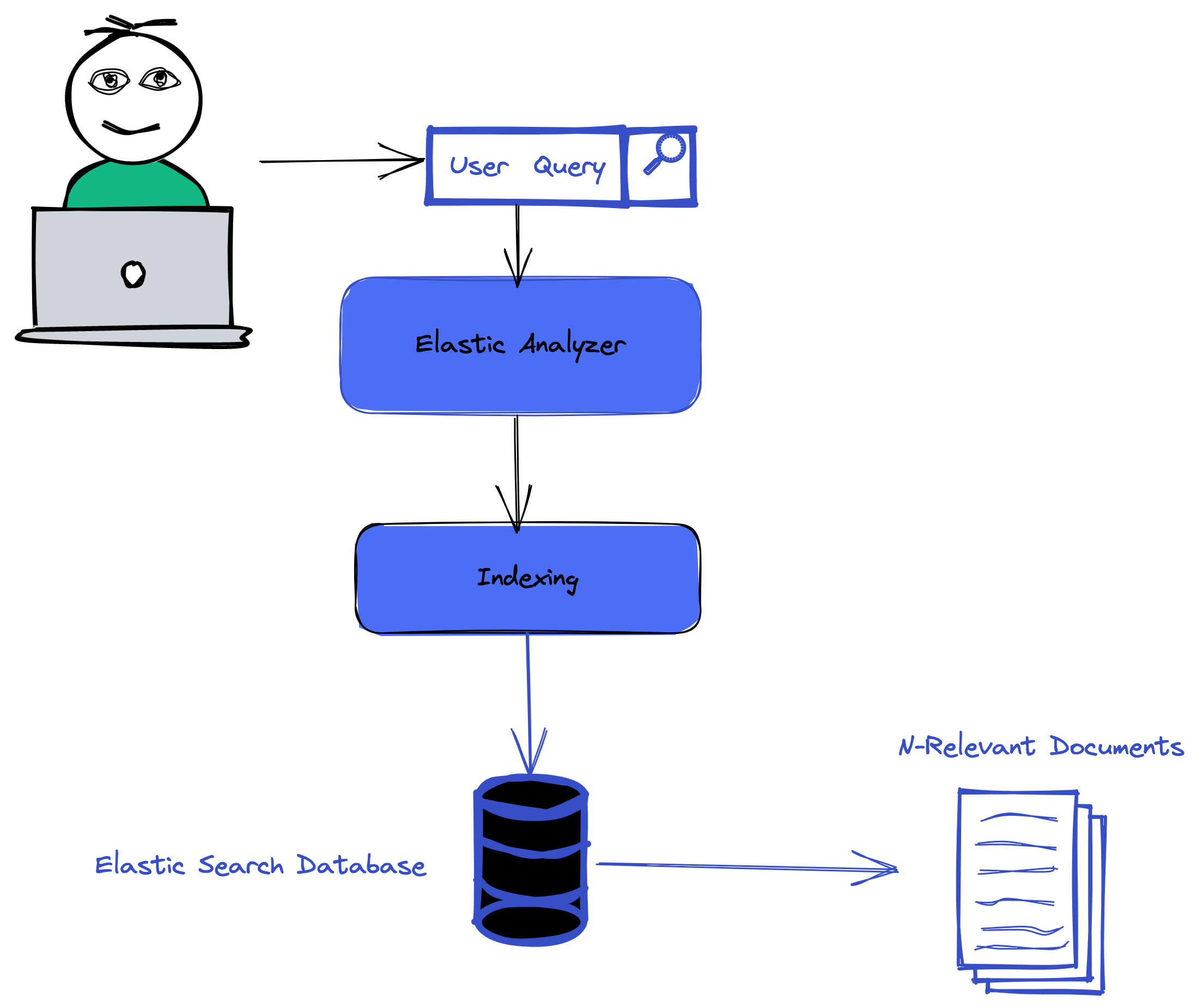
For the querying part of this tutorial we will run two differents types of queries , the text search and keyword search.
Text Search
The text search will perform direct search in the abstract of a search , it will try to find abstract that matches our text query. Let see how we can do that in elasticsearch.
from elasticsearch_dsl import Search
from elasticsearch_dsl import Q
connection = connections.get_connection()
search = Search(using=connection, index='covid19')
query = Q("multi_match", query=preprocess_text('are monkey responsible of ebola'), fields=['title', 'abstract'])
response = search.query(query).execute()
To process the response we will use the following :
for hit in response:
print(hit.meta.score, hit.title)
print(10 * '-')
45.725304 Potential for broad-scale transmission of Ebola virus disease during the West Africa crisis: lessons for the Global Health security agenda
----------
44.993748 One Health: Zoonoses in the Exotic Animal Practice
----------
42.261215 SARS-CoV-2 Aerosol Exhaled by Experimentally Infected Cynomolgus Monkeys
----------
40.333527 Controlling Ebola: what we can learn from China's 1911 battle against the pneumonic plague in Manchuria
----------
37.900352 Emergency response vaccines: lessons learned in response to communicable diseases.
----------
36.51764 Species-Specific Evolution of Ebola Virus during Replication in Human and Bat Cells
----------
36.44274 A pilot survey of the U.S. medical waste industry to determine training needs for safely handling highly infectious waste
----------
35.545017 Molecular Hydrogen as a Novel Antitumor Agent: Possible Mechanisms Underlying Gene Expression
----------
34.979034 Applying lessons from the Ebola vaccine experience for SARS-CoV-2 and other epidemic pathogens.
----------
33.92034 Sexual transmission and the probability of an end of the Ebola virus disease epidemic
----------
Keyword Search
For keyword search we will use the tf-idf vectorizer from the first serie to find the most important keywords in a search query and then find the articles that matches the given keywords.
def get_ifidf_for_words(text):
text = preprocess_text(text)
print(text)
tfidf_matrix= tf_idf_vectorizer.transform([text]).todense()
feature_names = tf_idf_vectorizer.get_feature_names()
feature_index = tfidf_matrix[0,:].nonzero()[1]
tfidf_scores = zip([feature_names[i] for i in feature_index], [tfidf_matrix[0, x] for x in feature_index])
tfidf_scores = dict(tfidf_scores)
return list(tfidf_scores.keys())
query_text = 'are monkey responsible of ebola'
def search_by_tf_idf(query):
"""
this perform the seach by query
"""
query_tfidf = get_ifidf_for_words(query)
search_query = Search(using=connections.get_connection(), index='covid19')
search_query = search_query.filter('terms', tags=query_tfidf)
results = search_query.execute()
return results
The above query will return the following results :
def print_results(results):
for hit in results:
print(hit.meta.score, hit.title)
print(10 * '-')
print(hit.abstract[:100])
print( 10 * '-')
By analyzing those results we can see that that the the tfd-idf approach yield better result that the text search
query_text = 'are gorila responsible of ebola'
results = search_by_tf_idf(query_text)
print_results(results)
gorila responsible ebola
0.0 Aquareovirus effects syncytiogenesis by using a novel member of the FAST protein family translated from a noncanonical translation start site.
----------
nonenvelope virus aquareoviruse orthoreoviruse unusual ability induce cellcell fusion syncy
----------
0.0 Neuropathological explanation of minimal COVID-19 infection rate in newborns, infants and children – a mystery so far. New insight into the role of Substance P
----------
sarscov novel coronavirus infection covid become global challenge affect elderly populatio
----------
query_text = "was ebola found in africa "
results = search_by_tf_idf(query_text)
print_results(results)
ebola find africa
0.0 Comparison of Outcomes in HIV-Positive and HIV-Negative Patients with COVID-19
----------
background south africa high prevalence hiv world date record high number case covid af
----------
0.0 Multimorbidity in South Africa: a systematic review of prevalence studies
----------
objective review prevalence study multimorbidity south africa identify prevalence estimate
----------
0.0 Riesgo de COVID-19 en españoles y migrantes de distintas zonas del mundo residentes en España en la primera oleada de la enfermedad
----------
introducción objetivos existen pocos estudio sobre el potencial papel de los orígene rac
----------
0.0 The State of the World’s Midwifery 2021 report: findings to drive global policy and practice
----------
third global state world midwifery report sowmy provide update evidence base sexual repr
----------
0.0 How Is the World Responding to the Novel Coronavirus Disease (COVID-19) Compared with the 2014 West African Ebola Epidemic? The Importance of China as a Player in the Global Economy
----------
article describe similarity difference response government international community current
----------
0.0 Clinical outcomes and cost-effectiveness of COVID-19 vaccination in South Africa
----------
low middleincome country implement covid vaccination strategy light vary uncertain vaccine
----------
0.0 While flattening the curve and raising the line, Africa should not forget street vending practices
----------
street vending practice common africa cater large portion continent population since iden
----------
query_text = "COVID-19: Is reinfection possible"
results = search_by_tf_idf(query_text)
print_results(results)
covid reinfection possible
0.0 What Is the Antibody Response and Role in Conferring Natural Immunity After SARS-CoV-2 Infection? Rapid, Living Practice Points From the American College of Physicians (Version 2).
----------
description scientific medical policy committee smpc american college physician acp develo
----------
0.0 SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma
----------
sarscov yv novel lineage coronavirus cause covid contain multiple mutation within two im
----------
0.0 Acute Inflammatory Painful Polyradiculoneuritis: An Uncommon Presentation Related to COVID-19.
----------
covid may associate neurologic complication patient covid may develop acute inflammatory
----------
0.0 The Semester COVID-19 Stole the Joy
----------
nursing faculty member describe covid pandemic disrupt nursing education
----------
0.0 Severe Acute Respiratory Syndrome Coronavirus 2 Reinfection Associates With Unstable Housing and Occurs in the Presence of Antibodies
----------
background factor associate severe acute respiratory coronavirus sarscov reinfection remain
----------
0.0 COVID-19: a confirmed case of reinfection in a nurse
----------
describe case yearold man report first confirm case covid reinfection campania region it
----------
Search using the python api
In the following section we will peform search using the python api itself and compare the results with the one return in the search by query section.
import json
from elasticsearch import Elasticsearch
es_client = Elasticsearch()
def run_query_loop():
""" Asks user to enter a query to search."""
while True:
try:
handle_query()
except KeyboardInterrupt:
break
return
def handle_query():
""" Searches the user query and finds the best matches using elasticsearch."""
query = input("Enter query: ")
query = preprocess_text(query)
search_start = time.time()
search = {"size": 10 ,"query": {"multi_match": {"query" : query, "fields": ["abstract", "title"]}}}
print(search)
response = es_client.search(index='covid19', body=json.dumps(search))
search_time = time.time() - search_start
print()
print("{} total hits.".format(response["hits"]["total"]["value"]))
print("search time: {:.2f} ms".format(search_time * 1000))
for hit in response["hits"]["hits"]:
print("id: {}, score: {}".format(hit["_id"], hit["_score"]))
print(hit["_source"]['title'])
print(10 * "==****")
# print(hit["_source"]['abstract'])
run_query_loop()
Enter query: COVID-19: Is reinfection possible
{'size': 10, 'query': {'multi_match': {'query': 'covid reinfection possible', 'fields': ['abstract', 'title']}}}
1087 total hits.
search time: 152.49 ms
id: 433, score: 49.442734
Phenylethanoid glycosides as a possible COVID-19 protease inhibitor: a virtual screening approach
==****==****==****==****==****==****==****==****==****==****
id: 622, score: 49.263004
Possible Adrenal Involvement in Long COVID Syndrome
==****==****==****==****==****==****==****==****==****==****
id: 31, score: 44.819622
Molecular Hydrogen as a Novel Antitumor Agent: Possible Mechanisms Underlying Gene Expression
==****==****==****==****==****==****==****==****==****==****
id: 369, score: 42.868736
Recombinant dimeric small immunoproteins neutralize transmissible gastroenteritis virus infectivity efficiently in vitro and confer passive immunity in vivo.
==****==****==****==****==****==****==****==****==****==****
id: 386, score: 38.94829
Germany’s next shutdown—Possible scenarios and outcomes
==****==****==****==****==****==****==****==****==****==****
id: 624, score: 35.84116
Factors associated with COVID-19 infections and mortality in Africa: a cross-sectional study using publicly available data
==****==****==****==****==****==****==****==****==****==****
id: 977, score: 35.332203
COVID-19, Obsessive-Compulsive Disorder and Invisible Life Forms that Threaten the Self
==****==****==****==****==****==****==****==****==****==****
id: 48, score: 33.553566
Ocular Surface Disease during dupilumab treatment in patients with atopic dermatitis, is it possible to prevent it?
==****==****==****==****==****==****==****==****==****==****
id: 530, score: 32.828606
The effect of COVID-19 on mental well-being in Switzerland: a cross-sectional survey of the adult Swiss general population
==****==****==****==****==****==****==****==****==****==****
id: 350, score: 32.144802
The Impact of COVID-19 Pandemic Infection in Patients Admitted to the Hospital for Reasons Other Than COVID-19 Infection.
==****==****==****==****==****==****==****==****==****==****
References
- Sarkar, D., 2022. A Practitioner’s Guide to Natural Language Processing (Part I) — Processing & Understanding Text. [online] Medium. Available at: https://towardsdatascience.com/a-practitioners-guide-to-natural-language-processing-part-i-processing-understanding-text-9f4abfd13e72 [Accessed 1 March 2022]
- Melanie Walsh, Introduction to Cultural Analytics & Python, Version 1 (2021).
- Allahyari, M., n.d. Text Search using TF-IDF and Elasticsearch, Machine Learning Tutorial. [online] Sci2lab.github.io. Available at: https://sci2lab.github.io/ml_tutorial/tfidf/ [Accessed 2 March 2022].
- Manning, C. D., Raghavan, P., & Schutze, H. (2008). Introduction to Information Retrieval. Cambridge University Press.
- SPARCK JONES, K. (1972), “A STATISTICAL INTERPRETATION OF TERM SPECIFICITY AND ITS APPLICATION IN RETRIEVAL”, Journal of Documentation, Vol. 28 No. 1, pp. 11-21. https://doi.org/10.1108/eb026526
Comments